Optimization and Application Development of Marine Cold-adapted DNA Polymerases - OptiZyme
Isothermal amplification methods are now increasingly being used for molecular detection of pathogens, and are considered an important contribution to improve both disease control and food safety analysis. The main goal of the project was to further develop preliminary characterized marine cold-adapted polymerases for improved efficiency and applicability in present and future isothermal amplification technologies. Efficient target amplification relies in many of the methods on the inherent strand-displacement activity (SDA) of the DNA polymerase used in the reaction setup. The term strand displacement describes the ability of the polymerase to displace downstream DNA encountered during synthesis and this particular property is exploited to perform strand separation of dsDNA. We have fully achieved our main goal to generate at least five candidate enzymes possessing significant better SDA than current state-of-the-art enzymes on the market. This was made possible through the generation of a molecular evolution library of the promising polymerase I from Psychrobacillus sp. characterized early within this project. Furthermore an efficient and specific mutant screening-platform has been established where mutants with increased SDA have been identified. The 3D-structure of the wild-type enzyme made it possible for us to map the mutated amino-acid residues in the structure. This will contribute to increase knowledge and understanding of the function of the enzyme what is highly important for further enzyme engineering through rational design.
 |
 |
|
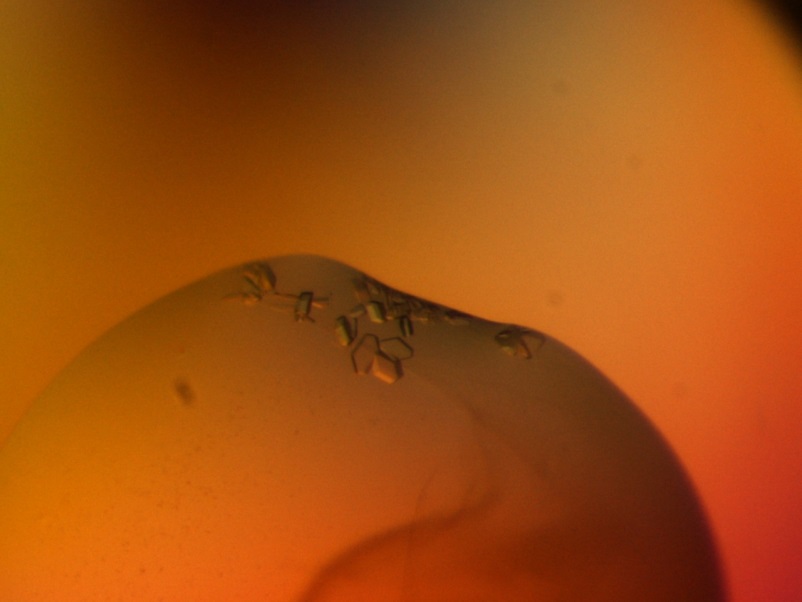
Crystals of marine cold-adapted polymerases: Starting point for structure determination and further improvement of enzymatic properties by rational design. |
|
Milestone results
- Patent filed - Marine DNA polymerases
- Commercial products – Isopol™DNA Polymerase
- Further fundings
- MDxPol - Marine DNA polymerases as tools for next generation Molecular Diagnostics (FORNY2020, NRC, 2016-2018)
- Marine DNA polymerases as engines for Single Cell Genomics (MABIT, 2017-2018)
- MarSynth - Marine DNA modifying enzymes for synthetic biology (BIOTEK2021, NRC, 2017-2020)
This project is an optimisation project funded by The Norwegian Research Council of Norway programme Biotechnology for innovation (BIOTEK2021) and ran at the Department of Chemistry at UiT - The Arctic University of Norway. The project period was October 2013 - October 2016.
For more information feel free to contact Atle N. Larsen.
Project team
Atle Noralf Larsen (project leader)
Yvonne Piotrowski (scientist)
Netsanet Gizaw Assefa (technician)
Man Kumari Gurung (technician)
Ronny Helland (scientist)