Kournoutis, A., Lamark, T., Johansen, T. and Abudu, Y.P. 2024. WDR83/MORG1 inhibits RRAG GTPase-MTORC1 signaling to facilitate basal autophagy. Autophagy, 7:1-2. doi: 10.1080/15548627.2024.2322457.
Abudu, Y.P., Kournoutis, A., Brenne, H.B., Lamark, T. and Johansen, T. 2024. MORG1 limits mTORC1 signaling by inhibiting Rag GTPases. Molecular Cell, 84(3):552-569.e11. doi: 10.1016/j.molcel.2023.11.023. Epub 2023 Dec 15.
Nähse, V., Raiborg, C., Tan, K.W., Mørk, S., Torgersen, M.L., Wenzel, E.M., Nager, M., Salo, V.T., Johansen, T., Ikonen, E., Schink, K.O. and Stenmark, H. 2023. ATPase activity of DFCP1 controls selective autophagy. Nature Communications, 14:4051. doi: 10.1038/s41467-023-39641-9.
Rogov, V.V., Nezis, I.P., Tsapras, P., Zhang, H., Dagdas, Y., Noda, N.N., Nkatogawa, H., Wirth, M., Mouilleron, S., McEwan, D.G., Behrends, C., Deretic, V., Elazar, Z., Tooze, S.A., Dikic, I., Lamark, T., and Johansen, T. 2023. Atg8 family proteins, LIR/AIM motifs and other interaction modes. Autophagy Reports 2, 2188523. doi: 0.1080/27694127.2023.2188523.
Rasmussen, N.L., Zhou, J., Olsvik, H., Kaeser-Pebernard, S., Lamark, T., Dengjel, J. and Johansen, T. 2023. The inflammation repressor TNIP1/ABIN is degraded by autophagy following TBK1 phosphorylation of its LIR. Autophagy Mar 9:1-3. doi: 10.1080/15548627.2023.2185013.
Zhou, J., Rasmussen, N.L., Olsvik, H.L., Akimov, V., Hu, Z., Evjen, G., Kaeser-Pebernard, S., Sankar, D.S., Roubaty, C., Verlhac, P., van de Beck, N., Reggiori, F., Abudu, Y.P., Blagoev, B., Lamark, T., Johansen, T. and Dengjel, J. 2023. TBK1 phosphorylation activates LIR-dependent degradation of the inflammation repressor TNIP1. J. Cell Biol. 222: e202108144. doi: 10.1083/jcb.202108144. Epub 2022 Dec 27.
McLaren Berge, A.K., Garcia-Garcia, J., Sjøttem, E., and Olsvik, H.L. 2023. The ubiquitin E3 ligase TRIM27 emerges as a new player in mitophagy. Autophagy Reports, 2, 2164089 doi: 10.1080/27694127.2022.2164089. ®
Olsvik, H.L. and Johansen, T. 2023. AlphaFold-multimer predicts ATG8 protein binding motifs crucial for autophagy research. PLoS Biol. 21, e3002002. doi: 10.1371/journal.pbio.3002002.
Kournoutis, A.. and Johansen, T. 2023. LC3B is a cofactor for LMX1B-mediated transcription of autophagy genes in dopaminergic neurons. J. Cell Biol. 222, e202303008. doi: 10.1083/jcb.202303008.
Wang, Y., Eapen, V.V., Kournoutis, A., Onorati, A., Li, X., Zhou, X., Cetinbas, M., Wang, L., Liu, J., Bretz, C., Zhou, Z., Sui, S.J.H., Saladi, S.V., Sadreyev, R.I., Adams, P.D., Kingston, R.E., Yue, Z., Johansen, T. and Dou, Z. 2022. Nuclear autophagy interactome unveils WSTF as a constitutive nuclear inhibitor of inflammation. bioRxiv 2022.10.04.510822; doi:10.1101/2022.10.04.510822
Garcia-Garcia, J., Berge, A.K.M., Overå, K.S., Larsen, K.B., Bhujabal, Z., Brech, A., Abudu, Y.P., Lamark, T., Johansen, T. and Sjøttem, E. 2022. TRIM27 is an autophagy substrate facilitating mitochondria clustering and mitophagy via phosphorylated TBK1. FEBS J. 290,1096-1116. doi:10.1111/febs.16628
Jia, J., Wang, F., Bhujabal, Z., Peters, R., Mudd, M., Duque, T., Allers, L., Javed, R., Salemi, M., Behrends, C., Phinney, B., Johansen, T. and Deretic, V. 2022. Membrane Atg8ylation, stress granule formation, and MTOR regulation during lysosomal damage. Autophagy doi: 10.1080/15548627.2022.2148900. Online ahead of print.®
Jia, J., Wang, F., Bhujabal, Z., Peters, R., Mudd, M., Duque, T., Allers, L., Javed, R., Salemi, M., Behrends, C,. Phinney, B., Johansen, T. and Deretic, V. 2022. Stress granules and mTOR are regulated by membrane atg8ylation during lysosomal damage. J. Cell Biol. 221, e202207091. doi: 10.1083/jcb.202207091
Rasmussen, N.L., Kournoutis, A., Lamark, T. and Johansen, T. 2022. NBR1: The archetypal selective autophagy receptor. J. Cell Biol. 221:e202208092. doi: 10.1083/jcb.202208092. ®
de la Riva Carrasco, R., Perez Pandolfo, S., Freire, S.S., Romero, N.M., Bhujabal, Z., Johansen, T., Wappner, P., and Melani, M. 2021. The immunophilin Zonda controls regulated exocytosis in endocrine and exocrine tissues. Traffic, 22, 111-122
Nthiga, T.M., Shrestha, B.K., S., Lamark, T., and Johansen, T. 2021. The soluble reticulophagy receptor CALCOCO1 is also a Golgiphagy receptor. Autophagy 17, 2051-2052. doi: 10.1080/15548627.2021.1940610. ®
Nthiga, T.M., Shrestha, B.K., S., Bruun, J.-A., Larsen, K.B., Lamark, T., and Johansen, T. 2021. Regulation of Golgi turnover by CALCOCO1-mediated selective autophagy. J. Cell Biol. 220:e202006128. doi: 10.1083/jcb.202006128.
Petridi, S., Jacomin, A-C., Bhujabal, Z., Johansen, T., and Nezis, I. 2021. Exploring selective autophagy in Drosophila: Methods to identify Atg8-interacting proteins. Methods Cell Biol. 165, 13-29. doi: 10.1016/bs.mcb.2020.10.008.
Klionsky, D……..Johansen, T……, And Tong, C.K. 2021. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 2021 Jan;17(1):1-382. doi: 10.1080/15548627.2020.1797280. Epub 2021 Feb 8. ®
Wirth, M., Mouilleron, S., Zhang, W., Sjøttem, E., Abudu, Y.P., Jain, A., Olsvik, H.L., Bruun, J-A., Razi, M., Jefferies, H.B.J., Lee, R., Joshi, D., O'Reilly, N., Johansen, T., and Tooze, S.A. 2021. Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity. J. Mol. Biol. 433, 166987. doi: 10.1016/j.jmb.2021.166987.
Garcia, J.G., Stange Overå, K., Khan, W., and Sjøttem, E. 2021. Generation of the short TRIM32 isoform is regulated by Lys 247 acetylation and a PEST sequence. PLoS One 16, e0251279. doi: 10.1371/journal.pone.0251279. eCollection 2021.
Abudu, Y.P., Shrestha, B.K., Zhang, W., Palara, A., Brenne, H.B., Larsen, K.B., Wolfson, D.L., Dumitriu, G., Øie, C.I., Ahluwalia, B.S., Levy, G., Behrends, C., Tooze, S.A., Mouilleron, S., Lamark, T. and Johansen, T. 2021. SAMM50 acts with p62 in piecemeal basal- and OXPHOS-induced mitophagy of SAM and MICOS components. J. Cell Biol. 220, e202009092. doi: 10.1083/jcb.202009092.
Abudu, Y.P., Mouilleron, S., Tooze, S.A., Lamark, T., and Johansen, T. 2021. SAMM50 is a receptor for basal piecemeal mitophagy and acts with SQSTM1/p62 in OXPHOS-induced mitophagy. Autophagy 17, 2656-2658. doi: 10.1080/15548627.2021.1953846.
Lamark, T., and Johansen, T. 2021. Mechanisms of Selective Autophagy. Ann. Rev. Cell Dev. Biol. 37, 143-169. doi: 10.1146/annurev-cellbio-120219-035530.
Claude-Taupin, A., Jia, J., Bhujabal, Z., Garfa-Traoré, M., Kumar, S., da Silva, G.P.D., Javed, R., Gu, Y., Allers, L., Peters, R., Wang, F., da Costa, L.J., Pallikkuth, S., Lidke, K.A., Mauthe, M., Verlhac, P., Uchiyama, Y., Salemi, M., Phinney, B., Tooze, S.A., Mari, M.C., Johansen, T., Reggiori, F., and Deretic, V. 2021. ATG9A protects the plasma membrane from programmed and incidental permeabilization. Nat. Cell Biol. 23, 846-858. doi: 10.1038/s41556-021-00706-w.
Aman, Y., Schmauck-Medina, T., Hansen, M., Morimoto, R.I., Simon, A.K., Bjedov, I., Palikaras, K. Simonsen, A., Johansen, T., Tavernarakis, N., Rubinsztein, D.C., Partridge, L., Kroemer, G., Labbadia, J. and Fang, E.F. 2021. Autophagy in healthy aging and disease. Nat. Aging, 1, 634–650. Doi: 10.1038/s43587-021-00098-4 ®
Klionsky, D.J., Petroni, G., Amaravadi, R.K., Baehrecke, E.H., Ballabio, A., Boya, P., Bravo-San Pedro, J.M., Cadwell, K., Cecconi, F., Choi, A.M.K., Choi, M.E., Chu, C.T., Codogno, P., Colombo, M.I., Cuervo, A.M., Deretic, V., Dikic, I., Elazar, Z., Eskelinen, E-L., Fimia, G.M., Gewirtz, D.A., Green, D.R., Hansen, M., Jäättelä, M., Johansen, T., Juhász, G., Karantza, V., Kraft, C., Kroemer, G., Ktistakis, N.T., Kumar,S., Lopez-Otin, C., Macleod, K.F., Madeo, F., Martinez, J.F., Meléndez, A., Mizushima, N., Münz, C., Penninger, J.M., Perera, R.M., Piacentini, M., Reggiori, F., Rubinsztein, D.C., Ryan, K.M., Sadoshima, J., Santambrogio, L., Scorrano, L., Simon, H-U., Simon, A.K., Simonsen, A., Stolz, A., Tavernarakis, N., Tooze, S.A., Yoshimori, T., Yuan, J., Yue, Z., Zhong, Q., Galluzzi, L., and Pietrocola, F. 2021. Autophagy in major human diseases. EMBO J. 40, e108863 doi: 10.15252/embj.2021108863 ®
Johansen, T., and Lamark, T. 2020. Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors. J. Mol. Biol. 432, 80-103.®
Shrestha, B.K., Skytte Rasmussen M., Abudu, Y.P., Bruun, J-A., Larsen, K.B., Alemu, E.A., Sjøttem, E., Lamark, T., and Johansen, T. 2020. NIMA-related kinase 9-mediated phosphorylation of the microtubule-associated LC3B protein at Thr-50 suppresses selective autophagy of p62/sequestosome 1. J Biol Chem. 295, 1240-1260.
Jakobi, A.J., Huber, S.T., Mortensen, S.A., Schultz, S.W., Palara, A., Kuhm, T., Shrestha, B.K., Lamark, T., Hagen, W.J.H., Wilmanns, M., Johansen, T., Brech, A., and Sachse, C. 2020. Structural basis of p62/SQSTM1 helical filaments and their role in cellular cargo uptake. Nature Commun. 11(1):440. doi: 10.1038/s41467-020-14343-8.
Mercado, N., Colley, T., Baker, J.R., Vuppussetty, C., Kono, Y., Clarke, C., Tooze, S., Johansen, T., and Barnes, P.J. 2020. Bicaudal D1 impairs autophagosome maturation in chronic obstructive pulmonary disease. FASEB Bioadv. 1, 688-705.
Jacomin, A.C., Petridi, S., Di Monaco, M., Bhujabal, Z., Jain, A., Mulakkal, N.C., Palara, A., Powell, E.L., Chung, B., Zampronio, C., Jones, A., Cameron, A., Johansen, T., and Nezis, I.P. 2020. Regulation of Expression of Autophagy Genes by Atg8a-Interacting Partners Sequoia, YL-1, and Sir2 in Drosophila. Cell Rep. 31(8):107695. doi: 10.1016/j.celrep.2020.107695.
Birgisdottir, Å.B, and Johansen, T. 2020. Autophagy and endocytosis - interconnections and interdependencies. J. Cell Sci. 133(10):jcs228114. doi: 10.1242/jcs.228114. ®
Nthiga, T.M., Shrestha, B.K., Sjøttem, E., Bruun, J-A., Bowitz Larsen, K., Bhujabal, Z., Lamark, T., and Johansen, T. 2020. CALCOCO1 acts with VAMP-associated proteins to mediate ER-phagy. EMBO J. 39(15):e103649. doi: 10.15252/embj.2019103649.
Nthiga, T.M., Shrestha, B.K., S., Lamark, T., and Johansen, T. 2020. CALCOCO1 is a soluble reticulophagy receptor. Autophagy 16, 1729-1731. doi: 10.1080/15548627.2020.1797289. Online ahead of print. ®
Xu, C., Wang, L., Fozouni, P., Evjen, G., Chandra, V., Jiang, J., Lu, C., Nicastri, M., Bretz, C., Winkler, J.D., Amaravadi, R., Garcia, B.A., Adams, P.D., Ott, M., Tong, W., Johansen, T., Dou, Z., and Berger, S.L. 2020. SIRT1 is downregulated by autophagy in senescence and ageing. Nature Cell Biol. 22, 1170-1179. doi: 10.1038/s41556-020-00579-5
Jacomin., A.C., Gohel, R., Hussain, Z., Varga, A., Maruzs, T., Eddison, M., Sica, M., Jain, A., Moffat, K.G., Johansen, T., Jenny, A., Juhasz, G., and Nezis, I.P. 2020. Degradation of arouser by endosomal microautophagy is essential for adaptation to starvation in Drosophila. Life Sci. Alliance, e202000965. doi: 10.26508/lsa.202000965.
Wang, L., Xu, C., Johansen, T., Berger, S.L., and Dou, Z. 2020. SIRT1 - a new mammalian substrate of nuclear autophagy. Autophagy 17, 593-595. doi: 10.1080/15548627.2020.1860541 ®
Jia J., Abudu, Y.P., Claude-Taupin, A., Gu, Y., Kumar, S., Choi, S.W., Peters, R., Mudd, M.H., Allers, L., Salemi, M., Phinney, B., Johansen, T., and Deretic, V. 2019. Galectins control MTOR and AMPK in response to lysosomal damage to induce autophagy. Autophagy 15, 169-171.
Olsvik, H.L., Svenning, S., Abudu, Y.P., Brech, A., Stenmark, H., Johansen, T., and Mejlvang, J. 2019. Endosomal microautophagy is an integrated part of the autophagic response to amino acid starvation. Autophagy 15, 182-183. ®
Rasmussen, M.S., Birgisdottir, Å.B., and Johansen, T. 2019. Use of Peptide Arrays for Identification and Characterization of LIR Motifs. Methods Mol. Biol. 1880, 149-161.
Birgisdottir, Å.B., Mouilleron, S., Bhujabal, Z., Wirth, M., Sjøttem, E., Evjen, G., Zhang, W., Lee, R., O'Reilly, N., Tooze, S.A., Lamark, T., and Johansen, T. 2019. Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs. Autophagy 15, 1333-1355.
Kumar, S., Gu, Y., Abudu, Y.P., Bruun, J.-A., Jain, A., Farzam, F., Mudd, M., Anonsen, J.H., Rusten, T.E., Kasof, G., Ktistakis, N., Lidke, K.A., Johansen, T., and Deretic, V. 2019. Phosphorylation of Syntaxin 17 by TBK1 Controls Autophagy Initiation. Dev. Cell 49, 130-144. e6. doi: 10.1016/j.devcel.2019.01.027.
Princely Abudu, Y., Pankiv, S., Mathai, B.J., Lystad, A. H., Bindesbøll, C., Brenne, H.B., Yoke, Wui Ng M., Thiede, B., Yamamoto, A., Mutugi Nthiga, T., Lamark, T., Esguerra, C.V., Johansen, T., and Simonsen, A. 2019. NIPSNAP1 and NIPSNAP2 Act as "Eat Me" Signals for Mitophagy. Dev. Cell 49, 509-525.e12. doi: 10.1016/j.devcel.2019.03.013.
Hoem, G., Bowitz Larsen, K., Øvervatn, A., Brech, A., Lamark, T., Sjøttem, E., and Johansen, T. 2019. The FMRpolyGlycine Protein Mediates Aggregate Formation and Toxicity Independent of the CGG mRNA Hairpin in a Cellular Model for FXTAS. Front. Genet. 10, 249. doi: 10.3389/fgene.2019.00249. eCollection 2019.
Johansen, T. 2019. Selective Autophagy: RNA Comes from the Vault to Regulate p62/SQSTM1. Curr. Biol. 29(8):R297-R299. doi: 10.1016/j.cub.2019.03.008. ®
Wirth, M., Zhang, W., Razi, M., Nyoni, L., Joshi, D., O'Reilly, N., Johansen, T., Tooze, S.A., and Mouilleron, S.V. 2019. Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins. Nat. Commun. 10(1):2055. doi: 10.1038/s41467-019-10059-6.
Abudu, Y.P., Pankiv, S., Mathai, B.J., Lamark, T., Johansen, T. and Simonsen, A. 2019. NIPSNAP1 and NIPSNAP2 act as "eat me" signals to allow sustained recruitment of autophagy receptors during mitophagy. Autophagy 15, 1845-1847. ®
Kehl, S.R., Soos, B.A., Saha, B., Choi, S.W., Herren, A.W., Johansen, T., and Mandell, M.A. 2019. TAK1 converts Sequestosome 1/p62 from an autophagy receptor to a signaling platform. EMBO Rep. Sep;20(9):e46238. doi: 10.15252/embr.201846238.
Pölönen, P., Jawahar Deen, A., Leinonen, H.M., Jyrkkänen, H.K., Kuosmanen, S., Mononen, M., Jain, A., Tuomainen, T., Pasonen-Seppänen, S., Hartikainen, J.M., Mannermaa, A., Nykter, M., Tavi, P., Johansen, T., Heinäniemi, M., and Levonen, A.L. 2019. Nrf2 and SQSTM1/p62 jointly contribute to mesenchymal transition and invasion in glioblastoma. Oncogene 38, 7473-7490.
Gu, Y., Princely Abudu, Y., Kumar, S., Bissa, B., Choi, S.W., Jia, J., Lazarou, M., Eskelinen, E.L., Johansen, T. and Deretic, V. 2019. Mammalian Atg8 proteins regulate lysosome and autolysosome biogenesis through SNAREs. EMBO J. 38:e101994. doi: 10.15252/embj.2019101994.
Overå, K. S., Garcia-Garcia, J., Bhujabal, Z., Jain, A., Øvervatn, A., Larsen, K.B., Deretic, V., Johansen, T., Lamark, T., and Sjøttem, E. 2019. TRIM32, but not its muscular dystrophy-associated mutant, positively regulates and is targeted to autophagic degradation by p62/SQSTM1. J Cell Sci. 132(23): jcs236596. doi: 10.1242/jcs.236596.
Schwob, A., Teruel, E., Dubuisson, L., Lormières. F., Verlhac, P., Abudu, Y.P., Gauthier, J., Naoumenko, M., Cloarec-Ung, F.M., Faure, M., Johansen, T., Dutartre, H., Mahieux, R., and Journo, C. 2019. SQSTM-1/p62 potentiates HTLV-1 Tax-mediated NF-κB activation through its ubiquitin binding function. Sci. Rep. 9(1):16014. doi: 10.1038/s41598-019-52408-x.
Hafrén, A., Üstün, S., Hochmuth, A., Svenning, S., Johansen, T., and Hofius, D. 2018. Turnip mosaic virus counteracts selective autophagy of the viral silencing suppressor HCpro. Plant Physiol., 176, 649–662
Fusco, C., Mandriani, B., Di Rienzo, M., Micale, L., Malerba, N., Cocciadiferro, D., Sjøttem, E., Augello, B., Squeo, G.M., Pellico, M.T., Jain, A., Johansen, T., Fimia, G.M., Merla, G. 2018. TRIM50 regulates Beclin 1 proautophagic activity. Biochim. Biophys. Acta, 1865, 908-919
Jia, J., Abudu, Y.P., Claude-Taupin, A., Gu, Y., Kumar, S., Choi, S.W., Peters, R., Mudd, M.H., Allers, L., Salemi, M., Phinney, B., Johansen, T., and Deretic, V. 2018. Galectins Control mTOR in Response to Endomembrane Damage. Mol. Cell, 70, 120-135
Mejlvang, J., Olsvik, H., Svenning, S., Bruun, J.-A., Abudu, Y.P., Bowitz Larsen, K., Brech, A., Hansen, T.E., Brenne, H., Hansen, T., Stenmark, H., and Johansen, T. 2018. Starvation induces rapid degradation of selective autophagy receptors by endosomal microautophagy. Journal of Cell Biology, 217, 3640-3655
Deretic V, Prossnitz E, Burge M, Campen MJ, Cannon J, Liu KJ, Sklar LA, Allers L, Garcia SA, Baehrecke EH, Behrends C, Cecconi F, Codogno P, Chen GC, Elazar Z, Eskelinen EL, Fourie B, Gozuacik D, Hong W, Hotamisligi G, Jäättelä M, Jo EK, Johansen T, Juhász G, Kimchi A, Ktistakis N, Kroemer G, MIzushima N, Münz C, Reggiori F, Rubinsztein D, Ryan K, Schroder K, Simonsen A, Tooze S, Vaccaro MI, Yoshimori T, Yu L, Zhang H, Klionsky DJ. 2018. Autophagy, Inflammation, and Metabolism (AIM) Center of Biomedical Research Excellence: supporting the next generation of autophagy researchers and fostering international collaborations. Autophagy 14, 925-929. ®
Katheder, N.S., Khezri, R., O’Farrell, F., Schultz, S.W., Jain, A., Rahman, M.M., Schink, K.O., Theodossiou, T.A., Johansen, T., Juhász, G., Bilder, D., Brech, A., Stenmark, H. and Rusten, T.E. 2017. Microenvironmental autophagy promotes tumour growth. Nature, 541, 417-420
Skytte Rasmussen, M., Mouilleron, S., Kumar Shrestha, B., Wirth, M., Lee, R., Bowitz Larsen, K., Abudu Princely, Y., O`Reilly, N., Sjøttem, E., Tooze, S.A., Lamark, T. and Johansen, T. 2017. ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of ATG8 family proteins. Autophagy, 13, 834-853
Johansen, T., Birgisdottir, Å.B., Huber, J., Kniss, A., Dötsch, V., Kirkin, V. and Rogov, V.V. 2017. Methods for Studying Interactions Between Atg8/LC3/GABARAP and LIR-Containing Proteins. Methods of Enzymol. 587, 143-169
Abreu, S., Kriegenburg, F., Gómez-Sánchez, R., Mari, M., Sánchez-Wandelmer, J.,Skytte Rasmussen, M., Soares Guimarães, R., Zens, B., Schuschnig, M., Hardenberg, R., Peter, M., Johansen, T., Kraft, C., Martens, S. and Reggiori, F. 2017. Conserved Atg8 recognition sites mediate Atg4 association with autophagosomal membranes and Atg8 deconjugation. EMBO Reports, 18, 765-780
Kimura, T., Jain, A., Choi, S.W., Mandell, M.A., Johansen, T. and Deretic, V. 2017. TRIM-directed selective autophagy regulates immune activation. Autophagy, 13, 989-990
Kimura, T., Jia, J., Claude-Taupin, A., Kumar, S., Choi, S.W., Gu, Y., Mudd, M., Dupont, N., Jiang, S., Peters, R., Farzam, F., Jain, A., Lidke, K.A., Adams, C.M., Johansen, T. and Deretic, V. 2017. Cellular and molecular mechanism for secretory autophagy. Autophagy, 13, 1084-1085
Kumar, S., Chauhan, S., Jain, A., Ponpuak, M., Choi, S.W., Mudd, M., Peters, R., Mandell, M.A., Johansen, T., and Deretic, V. 2017. Galectins and TRIMs directly interact and orchestrate autophagic response to endomembrane damage. Autophagy, 13,1086-1087
Bhujabal, Z., Birgisdottir, Å.B., Sjøttem, E., Brenne, H.B., Øvervatn A., Habisov, S., Kirkin, V., Lamark, T. and Johansen, T. 2017. FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Reports, 18, 947-961
Galluzzi, L., Baehrecke, E.H., Ballabio, A., Boya, P., Bravo-San Pedro, J.M., Cecconi, F., Choi, A.M., Chu, C.T., Codogno, P., Colombo, M.I., Cuervo, A.M., Debnath, J., Deretic, V., Dikic, I., Eskelinen, E.L., Fimia, G.M., Fulda, S., Gewirtz, D.A., Green, D.R., Hansen, M., Harper, J.W., Jäättelä, M., Johansen, T., Juhasz, G., Kimmelman, A.C., Kraft, C., Ktistakis, N.T., Kumar, S., Levine, B., Lopez-Otin, C., Madeo, F., Martens, S., Martinez, J., Melendez, A., Mizushima, N., Münz, C., Murphy, L.O., Penninger, J.M., Piacentini, M., Reggiori, F., Rubinsztein, D.C., Ryan, K.M., Santambrogio, L., Scorrano, L., Simon, A.K., Simon, H.U., Simonsen, A., Tavernarakis, N., Tooze, S.A., Yoshimori, T., Yuan, J., Yue, Z., Zhong, Q., and Kroemer, G. 2017. Molecular definitions of autophagy and related processes. EMBO J. 36, 1811-1836®
Melani, M., Valko, A., Romero, N.M., Aguilera, M.O., Acevedo, J.M., Bhujabal, Z., Perez-Perri, J., de la Riva-Carrasco, R.V., Katz, M.J., Sorianello, E., D'Alessio, C., Juhász, G., Johansen, T., Colombo, M.I., and Wappner, P. 2017. Zonda is a novel early component of the autophagy pathway in Drosophila. Mol. Biol. Cell, 28, 3070-3081
Tusco, R., Jacomin, A.C., Jain, A., Penman, B.S., Larsen, K.B., Johansen, T., and Nezis, I.P. 2017. Kenny mediates selective autophagic degradation of the IKK complex to control innate immune responses. Nature Commun., 8, 1264. doi:10.1038/s41467-017-01287-9
Lamark, T., Svenning, S., and Johansen, T. 2017. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem., 61, 609-624®
Puvirajesinghe, T.M., Bertucci, F., Jain, A., Scerbo, P., Belotti, E., Audebert, S., Sebbagh, M., Lopez, M., Brech, A., Finetti, P., Charafe-Jauffret, E., Chaffanet, M., Castellano, R., Restouin, A., Marchetto, S., Collette, Y., Gonçalvès, A., Macara, I., Birnbaum, D., Kodjabachian, L., Johansen, T. and Borg, J.P. 2016. Identification of p62/SQSTM1 as a component of non-canonical Wnt VANGL2-JNK signalling in breast cancer. Nature Communications 7, 10318. doi: 10.1038/ncomms10318
Klionsky, D.J., …. Johansen, T…….Lamark., T. and xxxx others. 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1-222
Kimura, T., Jain, A., Choi, S.W., Mandell, M.A., Johansen, T. and Deretic, V. 2016. TRIM-Directed Selective Autophagy Regulates Immune Activation. Autophagy, 13, 989-990
Habisov, S., Huber, J., Ichimura, Y., Akutsu, M., Rogova, N., Loehr, F., McEwan, D.G., Johansen, T., Dikic, I., Doetsch, V., Komatsu, M., Rogov, V. and Kirkin V. 2016. Structural and Functional Analysis of a Novel Interaction Motif within UFM1-activating Enzyme 5 (UBA5) Required for Binding to Ubiquitin-like Proteins and Ufmylation. J. Biol. Chem. 291, 9025-9041
Goode, A., Butler, K., Long, J., Cavey, J., Scott, D., Shaw, B., Sollenberger, J., Gell, C., Johansen, T., Oldham, N.J., Searle, M.S. and Layfield, R. 2016. Defective recognition of LC3B by mutant SQSTM1/p62 implicates impairment of autophagy as a pathogenic mechanism in ALS-FTLD. Autophagy, 12, 1094-1104
Hewitt, G., Carroll, B., Sarallah, R., Correia-Melo, C., Ogrodnik, M., Nelson, G., Otten, E.G., Manni, D., Antrobus, R., Morgan, B.A., von Zglinicki, T., Jurk, D., Seluanov, A., Gorbunova, V., Johansen, T., Passos, J.F. and Korolchuk V.I. 2016. SQSTM1/p62 mediates crosstalk between autophagy and the UPS in DNA repair. Autophagy, 12, 1917-1930
Mandell, M.A., Jain, A., Kumar, S., Castleman, M.J., Anwar, T., Eskelinen, E.L., Johansen, T., Prekeris, R. and Deretic, V. 2016. TRIM17 contributes to autophagy of midbodies while actively sparing other targets from degradation. J. Cell Sci. 129, 3562-3573
Chauan, S., Kumar, S., Jain, A., Ponpuak, M., Mudd, M.H., Kimura, T., Choi, S.W., Peters, R., Mandell, M., Bruun, J.-A., Johansen, T., and Deretic, V. 2016. TRIMs and Galectins Globally Cooperate and TRIM16 and Galectin-3 Co-direct Autophagy in Endomembrane Damage Homeostasis. Dev. Cell 39, 13-27
Kimura, T., Jia, J., Kumar, S., Choi, S.W., Gu, Y., Mudd, M., Dupont, N., Jiang, S., Peters, R., Farzam, F., Jain, A., Lidke, K.A., Adams, C.M., Johansen, T. and Deretic, V. 2016. Dedicated SNAREs and specialized TRIM cargo receptors mediate secretory autophagy. EMBO J. 36, 42-60
Borel, S., Robert-Hebmann, V., Alfaisal, J., Jain, A., Faure, M, Espert, L., Chaloin, L., Paillart, J.C., Johansen, T. and Biard-Piechaczyk, M. 2015. HIV-1 viral infectivity factor .with light chain 3 and inhibits autophagy. AIDS 29, 275–286
Bertrand M, Petit V, Jain A, Amsellem R, Johansen T, Larue L, Codogno P, Beau I. 2015. SQSTM1/p62 regulates the expression of junctional proteins through epithelial-mesenchymal transition factors. Cell Cycle 14, 364-374
Raiborg, C., Wenzel, E.M., Pedersen, N.M., Olsvik, H., Schink, K.O., Schultz, S.W., Vietri, M., Nisi, V., Bucci, C., Brech, A., Johansen, T. and Stenmark, H. 2015. Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature 520, 234-238
Bruun, J., Kolberg, M., Ahlquist, T.C., Røyrvik, E., Nome, T., Leithe, E., Lind, G.E., Merok, M.A., Rognum, T.O., Bjørkøy, G., Johansen, T., Lindblom, A., Sun, X.F., Svindland, A., Liestøl, K., Nesbakken, A., Skotheim, R.I. and Lothe, R.A. 2015. Regulator of chromosome condensation 2 identifies high-risk patients within both major phenotypes of colorectal cancer. Clin. Cancer Res. 21, 3759-3770
Ciuffa, R., Lamark, T., Tarafder, A.K., Guesdon, A., Rybina, S., Hagen, W.J., Johansen, T. and Sachse, C. 2015. The Selective Autophagy Receptor p62 Forms a Flexible Filamentous Helical Scaffold. Cell Rep. 11, 748-758
Johansen, T. and Sachse, C. 2015. The higher-order molecual rorganization of p62/SQSTM1. Oncotarget 6, 16796-16797
Jain, A., Rusten, T.E., Katheder, N., Elvenes, J., Bruun, J.A., Sjøttem, E., Lamark, T. and Johansen, T. 2015. p62/sequestosome-1, autophagy-related gene 8 and autophagy in Drosophila are regulated by Nuclear factor erythroid 2-related factor 2 (NRF2), independent of transcription factor TFEB. J. Biol. Chem. 290, 14945-14962
Kimura, T., Jain, A., Choi, S.W., Mandell, M.A., Schroder, K., Johansen, T. and Deretic, V. 2015. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J. Cell Biol. 210, 973-989
Olsvik, H.L., Lamark, T., Takagi, K., Bowitz Larsen, K., Evjen, G., Øvervatn, A., Mizushima, T. and Johansen, T. 2015. FYCO1 Contains a C-terminally Extended, LC3A/B-preferring LC3-Interacting Region (LIR) Motif Required for Efficient Maturation of Autophagosomes During Basal Autophagy. J. Biol. Chem. 290, 29361-29374
Dou, Z., Xu, C., Donahue, G., Shimi, T., Pan, J.A., Zhu, J., Ivanov, A., Capell, B.C., Drake, A.M., Shah. P.P., Catanzaro, J.M., Daniel Ricketts, M., Lamark, T, Adam, S.A., Marmorstein, R., Zong, W.X., Johansen, T., Goldman, R.D., Adams, P.D. and Berger, S.L. 2015. Autophagy mediates degradation of nuclear lamina. Nature 527, 105-109
Darvekar, S., Elvenes, J., Brenne, H. B., Johansen, T. and Sjøttem, E. 2014. SPBP is a Sulforaphane Induced Transcriptional Coactivator of NRF2 Regulating Expression of the Autophagy Receptor p62/SQSTM1. PLoS One 9,e85262. doi: 10.1371/journal.pone.0085262
Rubio, N., Verrax, J., Dewaele, M., Verfaillie, T., Johansen, T., Piette, J. and Agostinis, P. 2014. p38MAPK-regulated induction of p62 and NBR1 after photodynamic therapy promotes autophagic clearance of ubiquitin aggregates and reduces reactive oxygen species levels by supporting Nrf2–antioxidant signaling. Free Radical Biol. Med. 67, 292-303
Rogov, V., Dötsch, V., Johansen, T. and Kirkin, V. 2014. Interactions between Autophagy Receptors and Ubiquitin-like Proteins Form the Molecular Basis for Selective Autophagy. Mol. Cell 53,167-178
Kalvari, I., Tsompanis, S., Mulakkal, N.C., Osgood, R., Johansen, T., Nezis, I.P. and Promponas, V.J. 2014. iLIR: A web resource for prediction of Atg8-family interacting proteins. Autophagy 10, 913-925
Johansen, T. and Lamark, T. 2014. Selective autophagy goes exclusive. Nat Cell Biol. 16, 395-397
Mandell, M.A., Jain, A., Arko-Mensah, J., Chauhan, S., Kimura, T., Dinkins, C., Silvestri, G., Münch, J., Kirchhoff, F., Simonsen, A., Wei, Y., Levine, B., Johansen, T. and Deretic, V. 2014. TRIM Proteins Regulate Autophagy and Can Target Autophagic Substrates by Direct Recognition. Dev Cell. 30, 394-409
Mandell, M.A., Kimura, T., Jain, A., Johansen, T. and Deretic, V. 2014. TRIM proteins regulate autophagy: TRIM5 is a selective autophagy receptor mediating HIV-1 restriction. Autophagy 10, 2387-2388
Deosaran E., Larsen, K.B., Hua, R., Sargent, G., Wang, Y., Kim., S., Lamark, T., Jauregui, M., Law, K., Lippincott-Schwartz, J., Brech, A., Johansen, T. and Kim, P.K. 2013. NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci. 126, 939-952
Birgisdottir, A. B., Lamark, T. and Johansen, T. 2013. The LIR motif - crucial for selective autophagy. J. Cell Sci.126, 3237-3247
Svenning, S. and Johansen, T. 2013. Selective autophagy. Essays Biochem. 55, 79-92
Darvekar, S., Rekdal, C., Johansen, T. and Sjøttem, E. 2013. A Phylogenetic Study of SPBP and RAI1: Evolutionary Conservation of Chromatin Binding Modules. PLoS One 8,e78907. doi: 10.1371/journal.pone.0078907
Darvekar, S., Johnsen, S.S., Eriksen, A.B., Johansen, T. and Sjøttem, E. 2012. Identification of two independent nucleosome-binding domains in the transcriptional co-activator SPBP. Biochem. J. 442, 65-75
McKnight, N. C., Jeffries, H. B., Alemu, E. A., Saunders, R. E., Howell, M., Johansen, T. and Tooze, S. A. 2012. Genome-wide siRNA screen reveals amino acid starvation-induced autophagy requires SCOC and WAC. EMBO J. 31, 1931-1946
Sancho, A., Duran, J., García-España, A., Mauvezin, C., Alemu, E.A., Lamark, T., Macias, M.J., Desalle, R., Royo, M., Sala, D., Chicote, J.U., Palacín, M., Johansen, T., and Zorzano, A. 2012. DOR/Tp53inp2 and Tp53inp1 Constitute a Metazoan Gene Family Encoding Dual Regulators of Autophagy and Transcription. PLoS ONE 7, e34034.
Lamark, T. and Johansen, T. 2012. Aggrephagy: Selective Disposal of Protein Aggregates by Macroautophagy. Int. J. Cell Biol. 2012, 736905, 21 pages. doi:10.1155/2012/736905
Aichem, A., Kalveram, B. Spinnenhirn, V., Kluge, K., Catone, N., Johansen, T. and Groettrup, M. 2012. The proteomic analysis of endogenous FAT10 substrates identifies p62/SQSTM1 as a substrate of FAT10ylation. J. Cell Sci. 125, 4576-4585
Pilli, M., Arko-Mensah, J., Ponpuak, M., Roberts, E., Master, S., Mandell, M.A., Dupont, N., Ornatowski, W., Jiang, S., Bradfute, S.B., Bruun, J.A., Hansen, T.E., Johansen, T. and Deretic, V. 2012. TBK-1 Promotes Autophagy-Mediated Antimicrobial Defense by Controlling Autophagosome Maturation. Immunity, 37, 223-234
Klionsky, D.J., .....Johansen, T.,.......Lamark, T., ........ and 1267 others. 2012. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8, 445-544.
Kleine, H., Herrmann, A., Lamark, T., Forst, A.H., Verheugd, P., Lüscher-Firzlaff, J., Lippok, B., Feijs, K.L., Herzog, N., Kremmer, E., Johansen, T., Müller-Newen, G. and Luscher B. 2012. Dynamic subcellular localization of the mono-ADP-ribosyltransferase ARTD10 and interaction with the ubiquitin receptor p62. Cell Commun. Signal. 10:28 doi:10.1186/1478-811X-10-28
Alemu, E.A.,, Lamark, T., Torgersen, K.M., Birgisdottir, A.B., Bowitz Larsen, K., Jain, A., Olsvik, H., Øvervatn, A., Kirkin, V. and Johansen, T. 2012. ATG8 Family Proteins Act as Scaffolds for Assembly of the ULK Complex: Sequence Requirements for LC3-Interacting Region (LIR) Motifs. J. Biol. Chem. 287, 39275-39290
Johansen, T. and Lamark, T. 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy, 7, 279-296.
Hansen, T.E. and Johansen, T. 2011. Following autophagy step by step. BMC Biol. 9, 39. doi:10.1186/1741-7007-9-39
Mostowy, S., Sancho-Shimizu, V., Hamon, M.A., Simeone, R., Brosch, R., Johansen, T. and Cossart P.2011. p62 and NDP52 Proteins Target Intracytosolic Shigella and Listeria to Different Autophagy Pathways. J. Biol. Chem., 286, 26987-95
Svenning, S., Lamark, T., Krause, K. and Johansen, T. 2011. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy, 7, 993-1010
Elvenes, J., Thomassen, E.I.S., Sagen Johnsen, S., Kaino, K., Sjøttem, E. and Johansen, T. 2011. Pax6 Represses Androgen Receptor-Mediated Transactivation by Inhibiting Recruitment of the Coactivator SPBP. PLoS ONE 6, e24659.
Pankiv, S., Alemu, E.A., Brech, A., Bruun, J.A., Lamark, T., Øvervatn, A., Bjørkøy, G. and Johansen, T. 2010. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 188, 253-269
Pankiv, S., Lamark, T., Bruun, J.A., Øvervatn, A., Bjørkøy, G. and Johansen, T. 2010. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinitated proteins to promyelocytic leukemia bodies. J. Biol. Chem. 285, 5941-5953
Clausen, T. H., Lamark, T., Isakson, P., Finley, K., Bowitz Larsen, K., Brech, A., Øvervatn, A., Stenmark, H., Bjørkøy, G., Simonsen, A. and Johansen, T. 2010. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 6, 330-344
Filimonenko, M., Isakson, P., Finley, K.D. Anderson, M., Jeong, H., Melia, T.J., Bartlett, B.J., Myers, K.M., Birkeland, H.C., Lamark, T., Krainc, D., Brech, A., Stenmark, H., Simonsen, A., and Yamamoto, A. 2010. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol. Cell 38, 265-279. doi:10.1016/j.molcel.2010.04.007.
Lamark, T. and Johansen, T. 2010. Autophagy: links with the proteasome. Curr. Opin. Cell Biol. 22, 192-198
Ponpuak, M., Davis, A.S., Roberts, E.A., Delgado, M.A., Dinkins, C., Zhao, Z., Virgin, H.W. 4th, Kyei, G.B., Johansen, T., Vergne, I. and Deretic, V. 2010. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity 32, 329-341
Pankiv, S. and Johansen, T. 2010. FYCO1: Linking autophagosomes to microtubule plus end-directing molecular motors. Autophagy 6, 550-552
Jain, A., Lamark, T., Sjøttem, E., Bowitz Larsen, K., Awuh, J.A., Øvervatn, A., McMahon, M., Hayes, J.D. and Johansen, T. 2010. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem., 285, 22576–22591
Dikic, I. , Johansen, T. and Kirkin, V. 2010. Selective autophagy in cancer development and therapy. Cancer Res. 70, 3431-3434
Larsen, K.B., Lamark, T., Øvervatn, A., Harneshaug, I., Johansen, T. and Bjørkøy, G. 2010. A reporter cell system to monitor autophagy based on p62/SQSTM1. Autophagy 6, 784-793
Elvenes, J., Sjøttem, E., Holm, T., Bjørkøy, G. and Johansen, T. 2010. Pax6 localizes to chromatin-rich territories and displays a slow nuclear mobility altered by disease mutations. Cell. Mol. Life Sci. 67, 4079-4094
Nezis, I.P., Shravage, B.V., Sagona, A.P., Lamark, T., Bjørkøy, G., Johansen, T., Rusten, T.E., Brech, A., Baehrecke, E.H. and Stenmark, H. 2010. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J. Cell. Biol. 190, 523-531
Nezis, I.P., Shravage, B.V., Sagona, A.P., Johansen, T., Baehrecke, E.H. and Stenmark, H. 2010. Autophagy as a trigger for cell death: Autophagic degradation of inhibitor of apoptosis dBruce controls DNA fragmentation during late in nurse cells during late oogenesis in Drosophila. Autophagy, 6, 1214-1215.
Alemu, E.A., Sjøttem, E., Outzen, H., Larsen, K.B., Holm, T., Bjørkøy, G., Johansen, T. 2010. Transforming growth factor-β-inducible early response gene 1 is a novel substrate for atypical protein kinase Cs. Cell. Mol. Life Sci. 68, 1953-1968
Pursiheimo, J.P., Rantanen, K., Heikkinen, P.T., Johansen, T. and Jaakkola, P.M. 2009. Hypoxia-activated autophagy accelerates degradation of SQSTM1/p62. Oncogene 28, 334-344
Kirkin, V., Lamark, T., Sou, Y.S., Bjørkøy, G., Nunn, J.L., Bruun J-A., Shvets, E., McEwan, D.G., Clausen, T.H., Wild, P., Bilusic, I., Theurillat, J.P., Øvervatn, A., Ishii, T., Elazar, Z., Komatsu, M., Dikic, I. and Johansen, T. 2009. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505-516
Bjørkøy, G., Lamark, T., Pankiv, S., Øvervatn, A., Brech, A. and Johansen T. 2009. Monitoring autophagic degradation of p62/SQSTM1. Meth. Enzymol. 452, 181-197
Nezis, I. P., Lamark, T., Velentzas, A.D., Rusten, T.E., Bjørkøy, G., Johansen, T., Papassideri, I.S., Stravopodis, D.J., Margaritis, L.H., Stenmark, H. and Brech, A. 2009. Cell death during Drosophila melanogaster early oogenesis is mediated through autophagy. Autophagy 5, 298-302
Kirkin, V., Lamark, T., Johansen, T. and Dikic, I. 2009. NBR1 co-operates with p62 in selective autophagy of ubiquitinated targets. Autophagy 5, 732-733
Lamark, T., Kirkin, V., Dikic, I. and Johansen, T. 2009. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle 8, 1986-1990
Zheng, Y.T., Shahnazari, S., Brech, A., Lamark, T., Johansen, T. and Brumell, J.H. 2009. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J. Immunol. 183, 5909-5916
Klionsky, D.J. et al.,Johansen, T. et al., Zhu X., and Deter, R. L. 2008. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4, 151-175.
Grocott, T., Frost, V., Maillard, M., Johansen, T., Wheeler, G.N., Dawes, L.J., Wormstone, I.M. and Chantry, A. 2007. The MH1 domain of Smad3 interacts with Pax6 and
represses autoregulation of the Pax6 P1 promoter. Nucleic Acids Research, 35, 890-901.
Pankiv, S., Høyvarde Clausen, T., Lamark, T., Brech, A., Bruun, J-A.,Outzen, H., Øvervatn, Bjørkøy, G. and Johansen, T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131-24145.
Sjøttem, E., Rekdal, C., Svineng, G., Sagen Johansen, S., Klenow, H., Dale Uglehus, R. and Johansen, T. 2007. The ePHD protein SPBP interacts with TopBP1 and together they co-operate to stimulate Ets1-mediated transcription. Nucleic Acids Research 35, 6648-6662.
Bjørkøy, G., Lamark, T. and Johansen, T. 2006. p62/SQSTM1: A Missing Link between Protein Aggregates and the Autophagy Machinery. Autophagy, 2, 138-9.
Erdogan, E., Lamark, T., Stallings-Mann, M., Jamieson, L., Pellechia, M., Thompson, E.A., Johansen, T. and Fields, A.P. 2006. Aurothiomalate Inhibits Transformed Growth by Targeting the PB1 Domain of Protein Kinase C iota. J. Biol. Chem. 281, 28450-28459.
Gruber, F.X., Hjorth-Hansen, H, Mikkola, I., Stenke, L. and Johansen, T. 2006. A novel Bcr-Abl splice isoform is associated with the L248V mutation in CML patients with acquired resistance to imatinib. Leukemia, 20, 2057-2060.
Gruber, F.X., Lamark, T., Anonli, A., Sovershaev, M.A., Olsen, M. Gedde-Dahl, T., Hjort-Hansen, H., and Skogen, B. 2005. Selecting and deselecting imatinib-resistant clones: observations made by longitudinal, quantitative monitoring of mutated BCR-ABL. Leukemia 19, 2159-2165. doi: 10.1038/sj.leu.2403983.
Bruun, J-A., Thomassen, E.I.S., Kristiansen, K., Tylden, G., Holm, T., Mikkola, I., Bjørkøy, G. and Johansen, T. 2005. The third helix of the homeodomain of paired class homeodomain proteins acts as a recognition helix both for DNA and protein interactions. Nucleic Acids Res., 33, 2661-2675.
Bjørkøy, G., Lamark, T., Brech, A., Outzen, H., Perander, M., Øvervatn, A., Stenmark, H. and Johansen, T. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol., 171, 603-614.
Lamark, T., Perander, M., Outzen, H., Kristiansen, K., Øvervatn, A., Michaelsen, E., Bjørkøy, G. and Johansen, T. 2003. Interaction Codes within the Family of Mammalian Phox and Bem1p Domain-containing Proteins. J. Biol. Chem. 278, 34568-34581.
Lamark, T., Ingebrigtsen, M., Bjørnstad, C., Melkko, T., Mollnes, T.E., and Nielsen, E.W. 2001. Expression of active human C1 inhibitor serpin domian in Escherichia coli. Protein Expr. Purif. 22, 349-358. doi: 10.1006/prep.2001.1445.
Mikkola, I., Bruun, J-A., Holm, T. and Johansen, T. 2001. Superactivation of Pax6 mediated transactivation from paired domain-binding sites by DNA independent recruitment of different homeodomain proteins. J. Biol. Chem. 276, 4109-4118.
Perander, M., Bjørkøy, G. and Johansen, T. 2001. Nuclear import and export signals enable rapid nucleocytoplasmic shuttling of the atypical protein kinase C lambda. J. Biol. Chem. 276, 13015-13024.
Rekdal, C., Sjøttem, E. and Johansen, T. 2000. The nuclear factor SPBP contains different functional domains and stimulates the activity of various transcriptional activators. J. Biol. Chem. 275, 40288-40300.
Mikkola, I., Bruun, J-A., Bjørkøy, G., Holm, T. and Johansen, T. 1999. Phosphorylation of the transactivation domain of Pax6 by extracellular signal-regulated kinase and p38 mitogen-activated protein kinase. J. Biol. Chem. 274, 15115-15126.
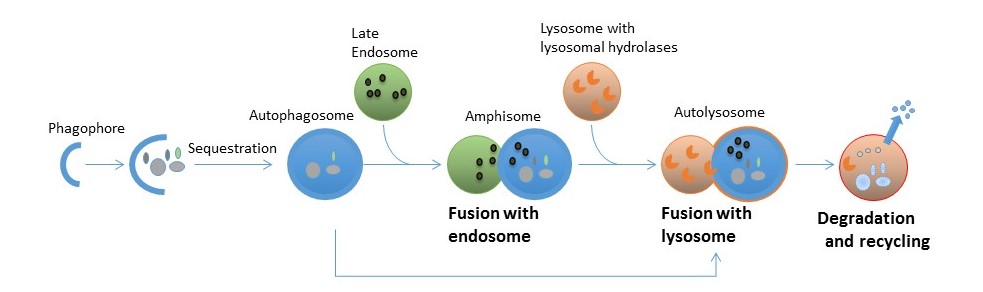




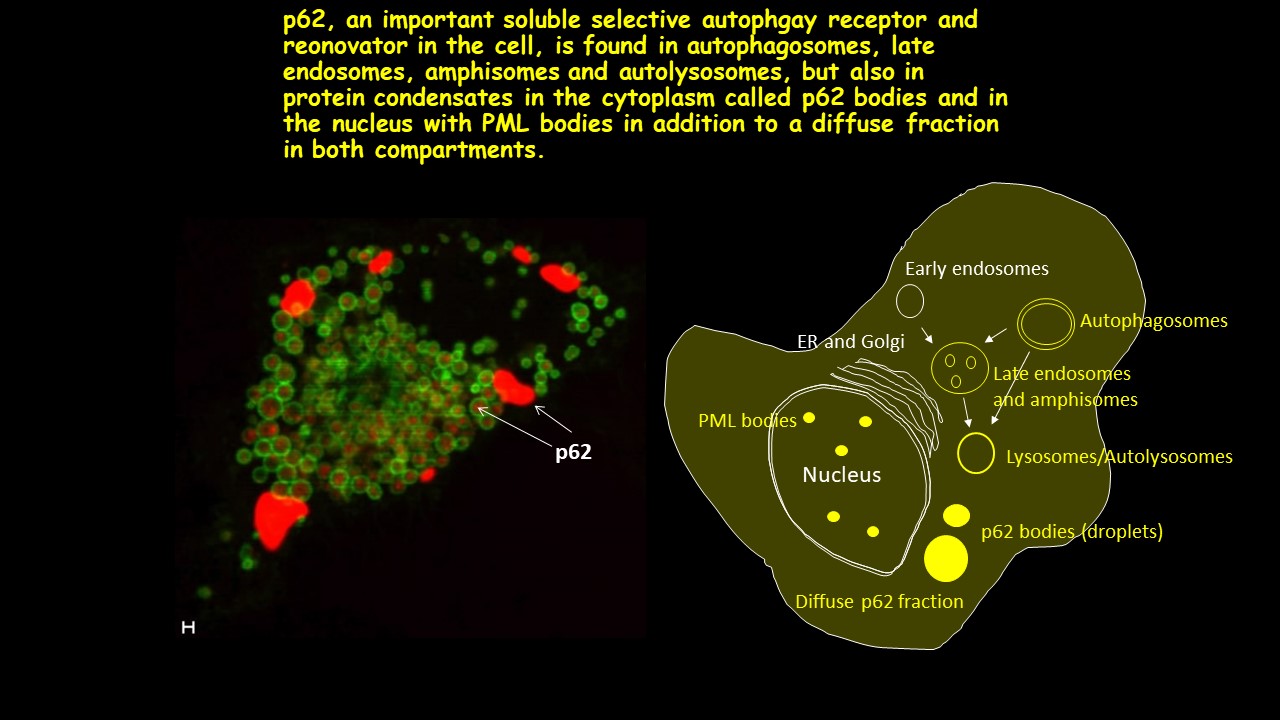
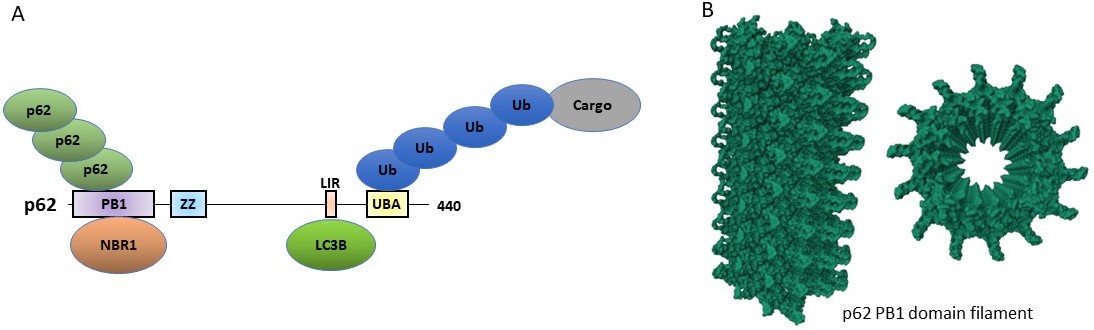
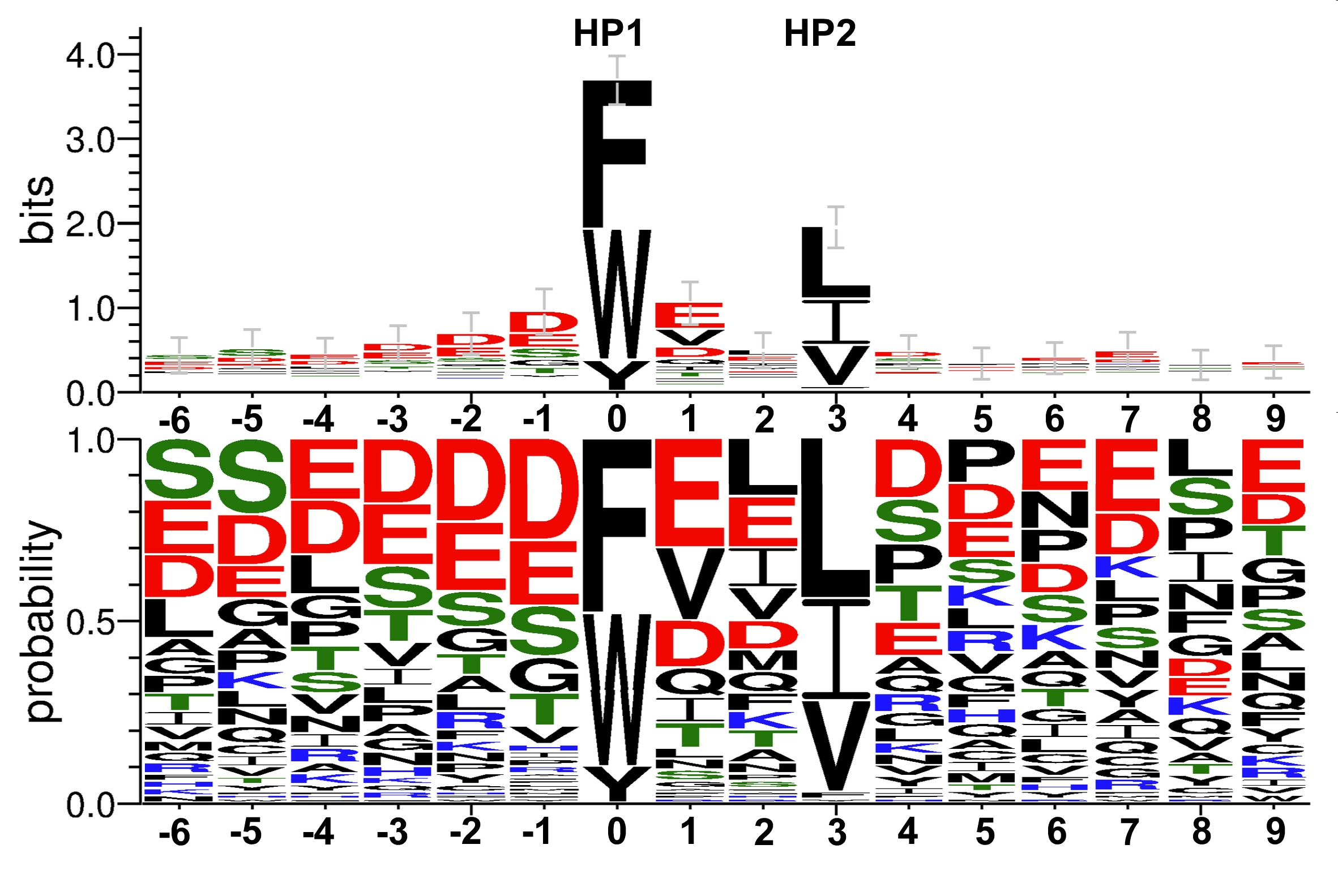 LIR motifs (100 different) compiled from proteins binding to Atg8/LC3/GABARAP proteins. For more information see
LIR motifs (100 different) compiled from proteins binding to Atg8/LC3/GABARAP proteins. For more information see